Motivations and principle
The purpose of stoic (supervised learning to inform clustering) is to extract meaningful associations between two data modalities. In essence, observations are clustered in the space of the main modality (e.g. gene/CRE expression), informed by the second one (e.g. their DNA sequence). This allows to simultaneously recover associations between
- groups of co-varying observations in the 1st dataset (e.g co-expression clusters) and
- their specific features in the 2nd dataset (e.g which DNA sequence features are predictive of a given co-expression cluster).
This procedures improves upon two-step approaches (i.e clustering observations in the main dataset, followed by an enrichment analysis of the second type of features), the results of which strongly rely on the upstream clustering strategy and parameters tuning. Furthermore, stoic does not make the assumption of a perfect correspondence between the two data sources.
The core novelty of stoic is that it performs the unsupervised clustering task by leveraging a supervised framework: it does so through an exploration of the main modality (e.g gene expression) space where clusters are iteratively formed by maximizing the performance of a classification model predicting cluster membership based on the orthogonal features. Each cluster thus corresponds to a supervised classifier, trained to discriminate the cluster from other observations using the second modality as predictive variables. From such models, feature importance estimates can be extracted and used to interpret clusters.
While we developed and used stoic in the context of systems & regulatory genomics, we believe that it could provide insights into many other fields and large datasets.
Using stoic
Installation
stoic can be installed directly from gitlab:
library(remotes) # remotes can be installed classically from the CRAN
install_gitlab(c("oceane.cssn/extendedforest", "oceane.cssn/stoic"))Input for stoic
Two data matrices are needed to run stoic:
The main data matrix, giving for each observations values in a set of samples. We recommend using several hundreds or even thousands of observations in
stoic, measured in several tenths or hundreds of samples.The guide data matrix, that gives for each observation a set of orthogonal features intended to inform the clustering in the main data. We tested
stoicfor several tenths or hundreds of guide features.
Usage and output example
Here is how to use stoic on its demonstration dataset, i.e finding the DNA features underlying the transcriptional activity of regulatory regions during neuron differentiation:
library(stoic)
# data upload and preparation
data("neuron_diff")
data <- stoic_data(main_data = neuron_diff$main_data,
guide_data = neuron_diff$guide_data,
sample_order = neuron_diff$sample_order, seed = 666)
# running stoic
stoic_results <- stoic_clustering(data, nstart = 2, n_cores = 4, n_trees = 500, seed = 666)
# Plotting results
draw_tsne(data, stoic_results) +
draw_profiles(data, stoic_results)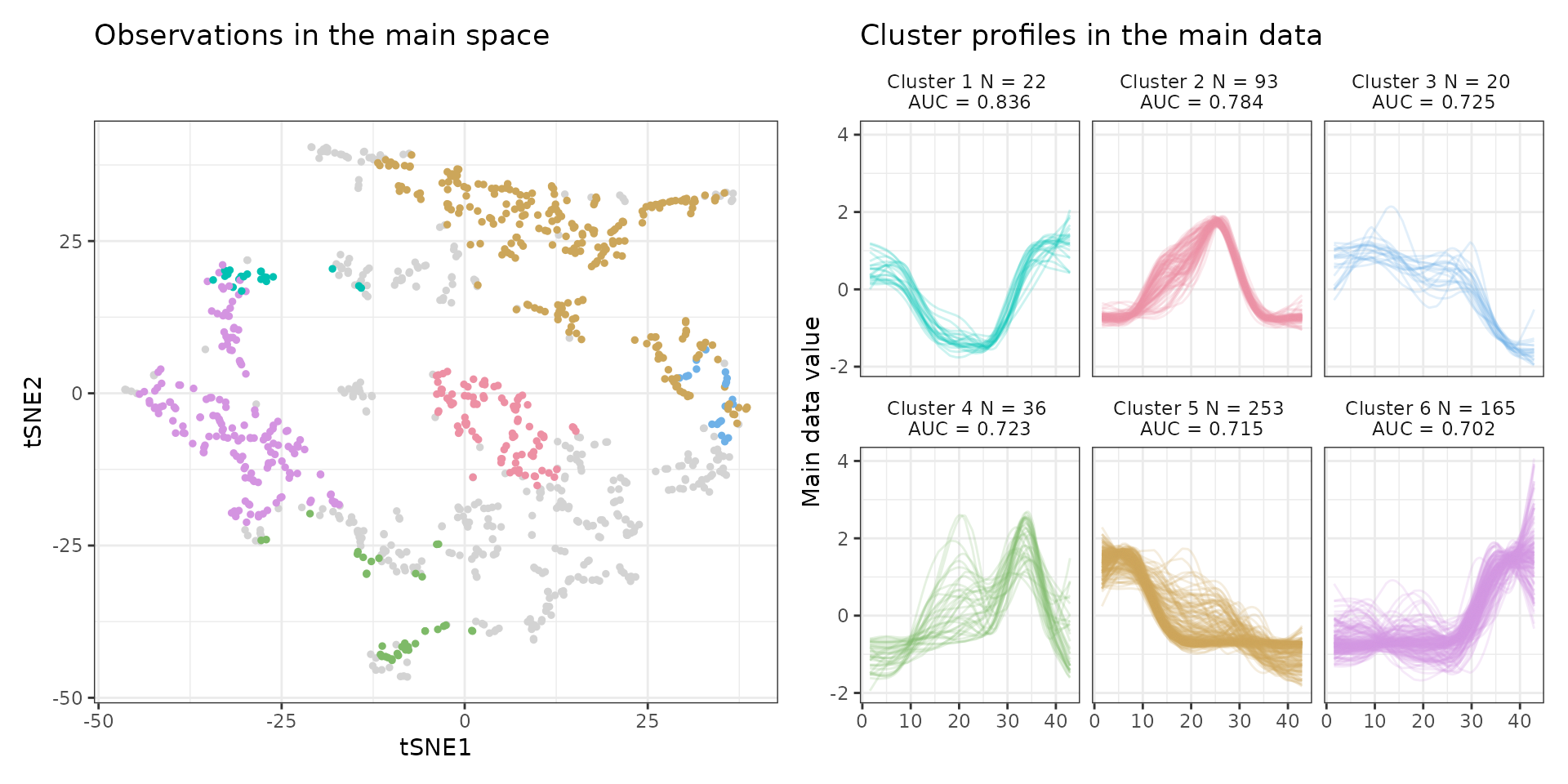
Before setting the seed, ensure that n_start is sufficiently large and provides stable results between different runs without fixed seeds.
Also, if the number of observations per clusters is too low, you might want to increase the imbalance_limit threshold (to 0.1 for example, instead of the default 0.025 appropriate to large numbers of observations).
This shows the clusters of co-expressed regulatory sequences, guided by nucleotidic composition. The sequence-level features associated with a co-expression cluster (number 2 here) can be observed with:
i = 2; draw_tsne(data, stoic_results, cluster = i) +
draw_profiles(data, stoic_results, cluster = i) +
draw_features(data, stoic_results, cluster = i)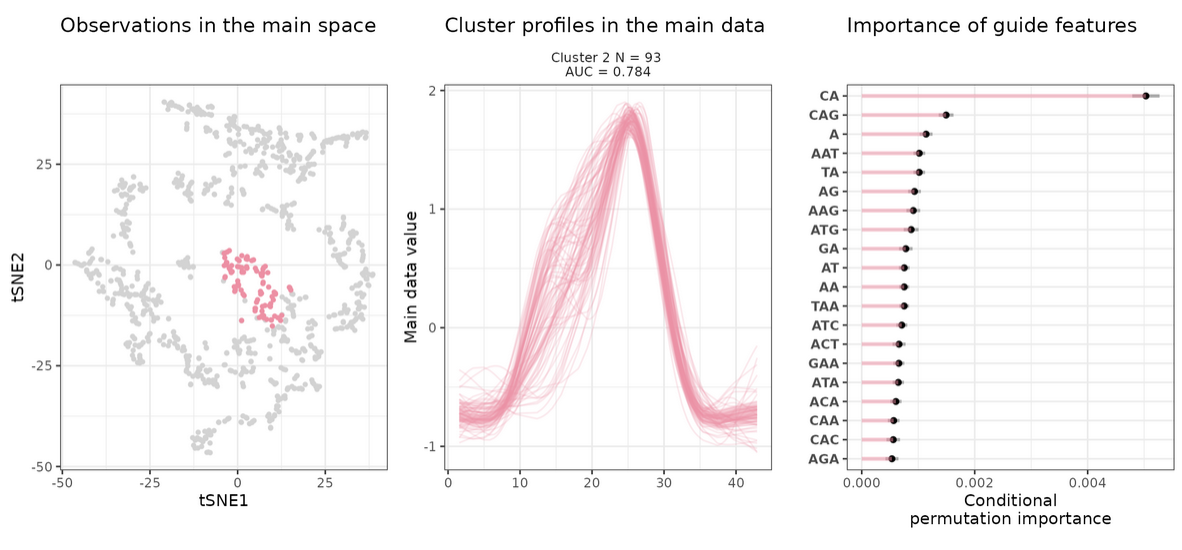
You can browse the corresponding Article for more details about the stoic functions and this use case.
A second vignette (CO2 gradient case study) demonstrates the use of stoic on a plant biology case study.
Reference
The full package documentation can be found in the Gitlab Page.
License
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.
Authors : Océane Cassan, Charles Lecellier, Laurent Bréhélin
